CHELTER - Role of intra-Clonal HEterogeneity and Leukemic environment in ThErapy Resistance of chronic leukemias
Published on January 11, 2022 – Updated on January 11, 2022
Our goal is to understand the role of molecular abnormalities (methylation, telomere dynamics, and metabolome) other than classical genetic drivers on the course of chronic myeloid leukaemia (CML) and chronic lymphocytic leukaemia (CLL) and their response to targeted therapies in order to identify new biomarkers or therapeutic strategies.
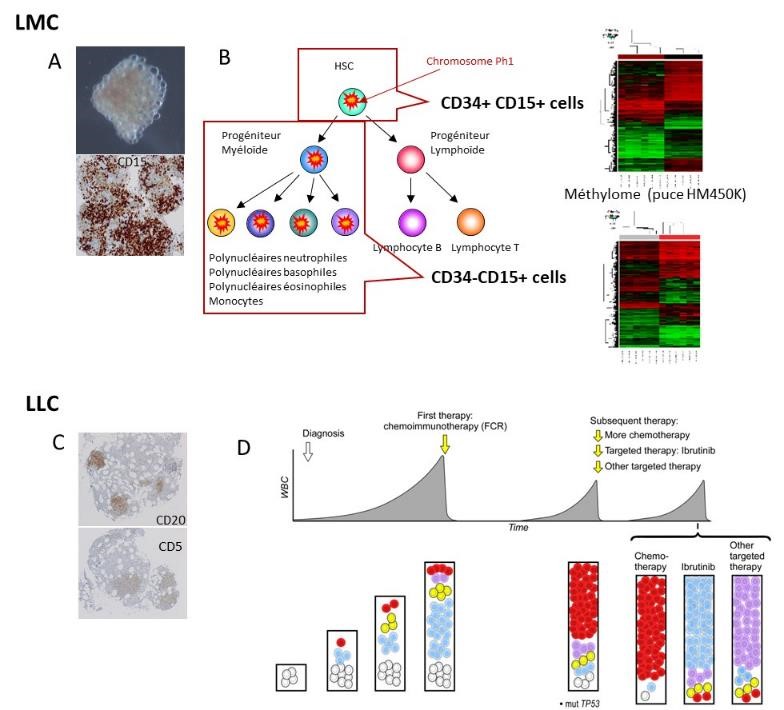
A and C: Identification of CML and CLL cells in medullary hematoma. B: Demonstration of intra-clonal epigenetic heterogeneity (DNA methylation) of the CML clone at diagnosis. D: Subclonal genetic evolution of CLL
These chronic blood malignancies concern people with a median age of 50-65 years and their management has recently been improved by the development of particularly effective targeted therapies. However, the therapeutic response is heterogeneous and disease cure is obtained only in a minority of patients. Therefore, it is essential to understand the mechanisms of resistance/sensitivity to targeted therapies according to the individual patient characteristics.
Our strategy is based on the study of primary malignant cells (close to the in vivo conditions) from two chronic leukaemia types: CML and CLL.
In CML, the different cell populations can be identified by flow cytometry. We could demonstrate the variability in tyrosine kinase inhibitor (TKI) targeting within each patient’s clone and among patients. As all cells in the CML clone have the same genetic driver (the BCR::ABL1 chimeric gene) targeted by TKIs, other mechanisms must be involved, especially epigenetic alterations. Indeed, we showed the intra-clonal epigenetic (DNA methylation) heterogeneity, with a specific profile of immature leukemic cells. A project is under way to identify methylation marks linked to resistance or sensitivity to TKIs (EPIK protocol), and to study non-malignant haematopoiesis in order to better understand the relationship between the patient's genetic/epigenetic background, telomere dynamics, and CML management. Our preliminary results also show that in CML cells, biological ageing (DNA methylation clock) is altered. Understanding the relationship between biological age, individual medical frailty and their influence on CML management is currently a major research axis of the CML subgroup.
In CLL, we demonstrated by tracking gene abnormalities identified by NGS that the CLL clone can adapt to each targeted therapy.
CLL cells are characterized by their dependence on the anti-apoptotic protein BCL2, a well-recognized therapeutic target. Venetoclax is a new BCL2 inhibitor currently used in patients who relapsed and soon as first-line treatment. However, relapse remains the rule, sometimes after a deep and prolonged response. Our team is interested in the mechanisms regulating resistance to venetoclax. Our first approach, based on the genome-wide screening and analysis of resistant cells from patients with CLL, revealed the involvement of the anti-apoptotic protein MCL1 and energy metabolism. Our current research projects are focused on i) metabolomic analyses to identify the pathways driving oxidative phosphorylation in venetoclax-resistant CLL cells, and ii) the characterization of a national cohort of patients with CLL resistant to venetoclax.
Lastly, CLL cells can resist treatment also by undergoing transformation into aggressive lymphoma (Richter syndrome). We are investigating the mutational landscape and gene expression changes of this syndrome by comparing the profile of CLL and Richter syndrome cells in the same patient.
Other GCCA partners
Lab
Hématologie Biologique, CHU Estaing
EA 7453 CHELTER
1 place Lucie et Raymond Aubrac
63003 Clermont-Ferrand Cedex 1